Our laboratory investigates the mechanisms of entry and fusion of enveloped viruses, including HIV-1, Influenza virus, arenaviruses, coronaviruses and other human pathogens. We use functional assays and real-time virus tracking to delineate the entry and fusion steps of infection of diverse viruses. We visualize the release of single viral content into the cytoplasm and subsequent viral core trafficking and uncoating that culminate in infection. We are also interested in understanding the molecular interactions involved in HIV-1 docking and the nuclear pore complex and nuclear import. A major focus in the lab is on understanding the mechanisms of antiviral activity of host restriction factors that target virus entry/fusion, such as IFITM3 and SERINC5. In addition to live cell imaging, we are also employing super-resolution microscopy and correlative light-electron microscopy (CLEM) techniques to delineate key structural intermediate steps of virus entry.
https://youtu.be/b-tZWZ4vnlI
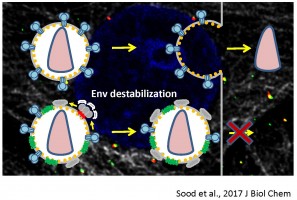
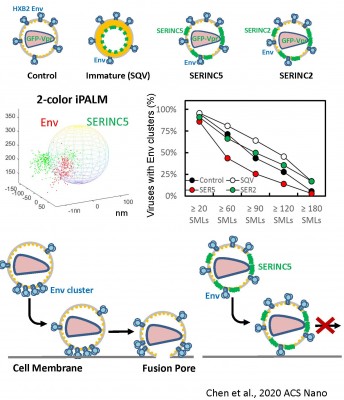
In the absence of HIV/SIV Nef expression, SERINC5 incorporates into budding virions and inhibits viral fusion, but the mechanism of restriction remains unclear. Envelope glycoproteins (Env) from different HIV-1 isolates exhibit a broad range of sensitivity to SERINC5. We have shown that SERINC5 incorporation alters Env conformation and sensitizes Env to neutralizing antibodies and fusion inhibitors. We also found that SERINC5 accelerates spontaneous inactivation of Env on virions (Figure 1). Using 2D- and 3D-superresolution imaging, we have recently demonstrated that SERINC5, but not its inactive analog SERINC2, disrupts functional Env clusters on mature HIV-1 particles (Figure 2). Thus, SERINC5 appears to both destabilize HIV-1 Env, thus accelerating its inactivation, and reduce infectivity through disrupting Env clusters that are the likely sites of virus-cell fusion.
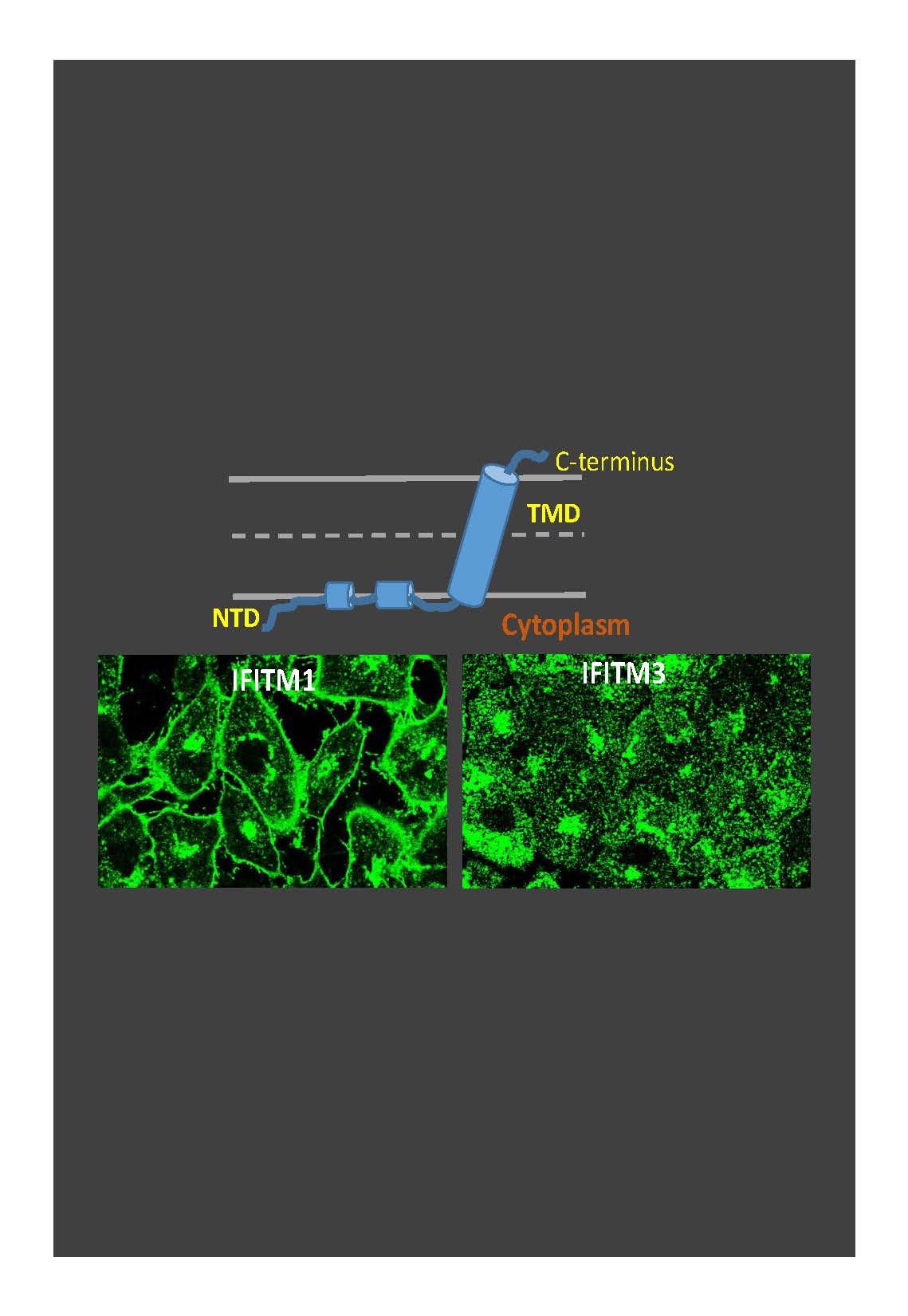
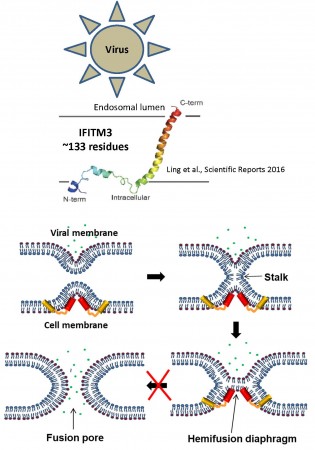
IFITMs (Figure 3) inhibit infection of diverse enveloped viruses, including the Influenza A virus, West Nile, Ebola and others. Using single virus imaging, we have shown that IFITM3 concentrates at the sites of Influenza virus entry and allows lipid mixing but abrogates the release of viral content into the cytoplasm (Figure 3, Movies 1 and 2). We also found that arenaviruses, which are resistant to IFITM restriction, traffic and fuse with endosomal compartments devoid of IFITM3. Our unpublished data imply that IFITM3 imposes negative curvature and increases rigidity of the cytoplasmic leaflet of a target membrane, thereby disfavoring the formation of fusion pores.
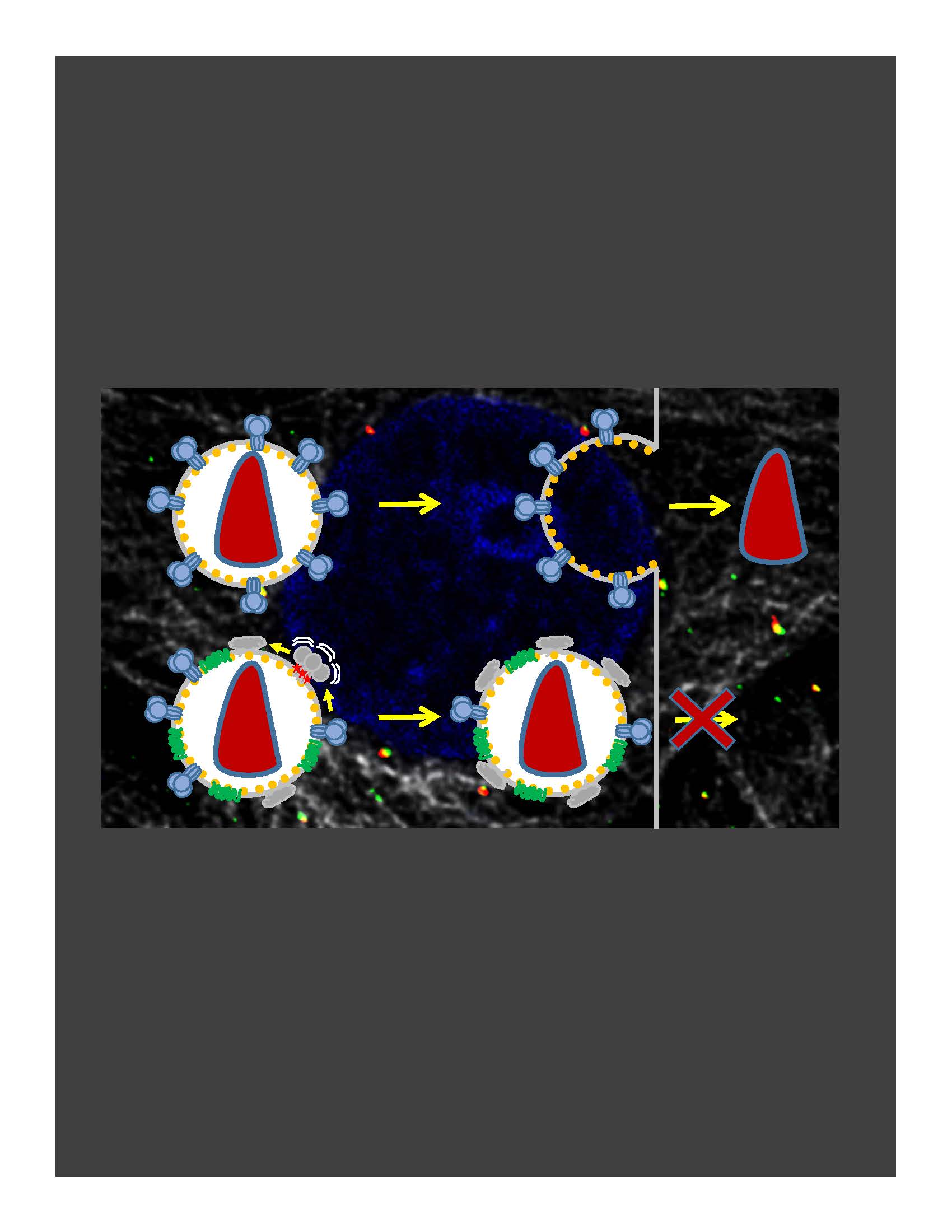
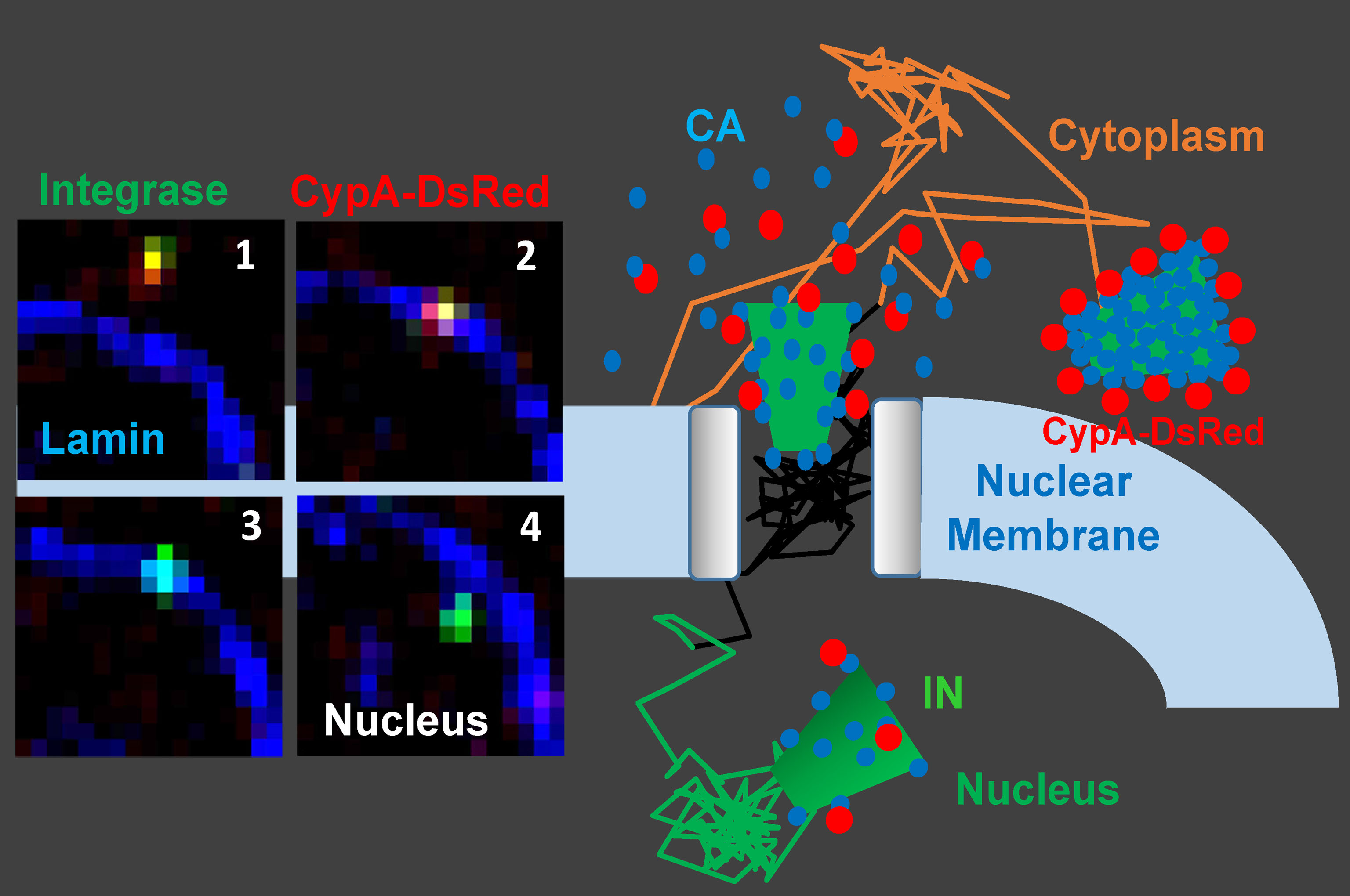
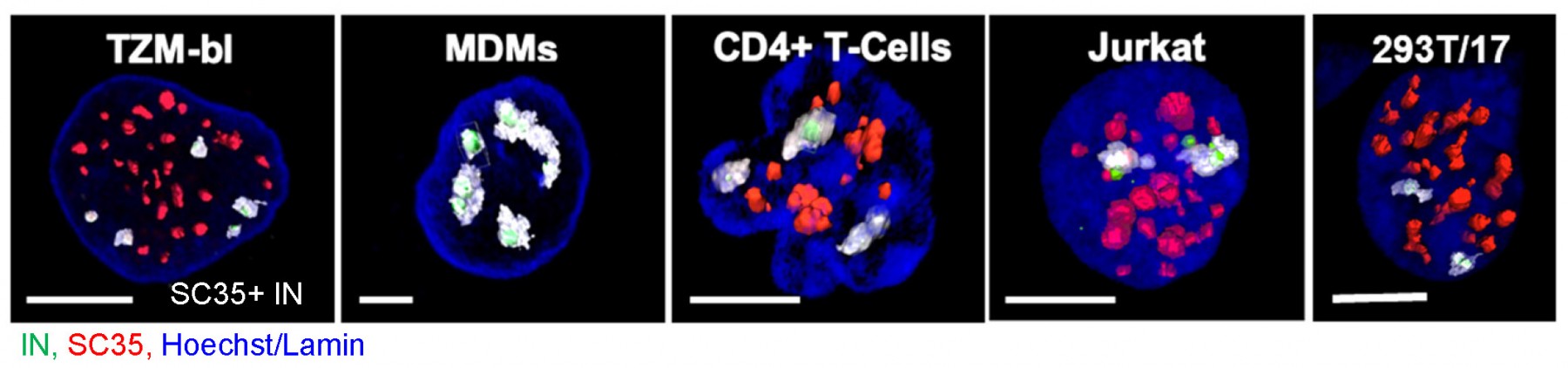
Disassembly of the cone-shaped HIV-1 capsid/core (uncoating) after virus-cell fusion is a prerequisite for establishing infection. We have visualized HIV-1 uncoating using a fluorescently tagged oligomeric form of a capsid-binding host protein cyclophilin A (CypA-DsRed). CypA-DsRed is specifically packaged into virions and does not compromise infectivity. Through the use of this HIV-1 capsid protein (CA) marker and long-term live cell imaging, we tracked single virus cores and showed that productive uncoating that leads to infection (reporter GFP expression) occurs at the nuclear pore (Figure 4 and Movie 3). We found that CA shell protects viral complexes from degradation in the cytoplasm and is required for docking at the nuclear pore. More recently, we investigated HIV-1 trafficking, uncoating and nuclear entry in primary human macrophages and found that viral pre-integration complexes are transported to nuclear speckles in macrophages and other cell types where they integrate into speckle-associated genomic domains, SPADs.

Correlated light-electron microscopy (CLEM)
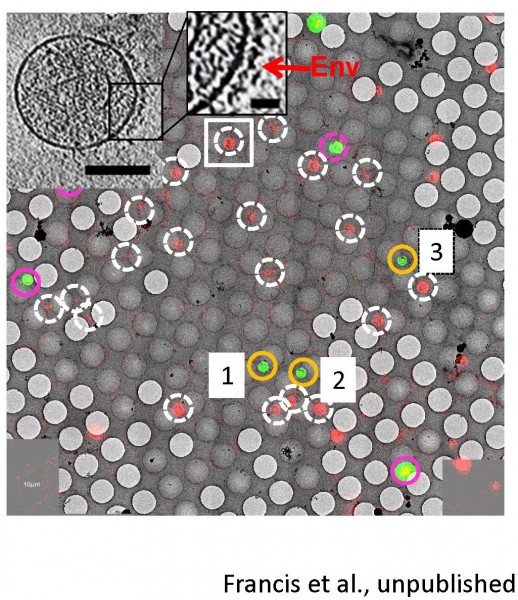
Ashwanth Francis in the lab is leading the efforts to implement correlated single virus fluorescence microscopy/cryo-electron tomography to gain structural insights into viral fusion and subsequent HIV-1 uncoating intermediates. Proof-of-concept CLEM images and a single HIV-1 particle tomogram are shown in Figure 6.
Measuring early virus-cell interactions using DNA force probe
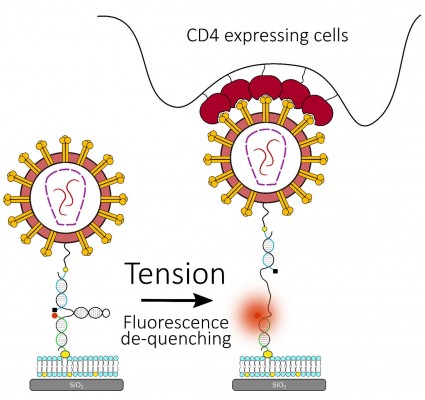
To understand the role of membrane tension in regulating HIV-1 fusion, we are developing a platform using Molecular Tension Force Microscopy (MTFM), in collaboration with Khalid Salaita (Emory). MTFM utilizes a surface-immobilized DNA hairpin-based probe, with a fluorophore-quencher pair that is designed to unfold upon the application of force. When the applied force exceeds the unfolding force of the hairpin, the spatial separation of the quencher-fluorophore pair leads to a strong enhancement of the fluorescence signal (Figure 7). If successful, this project will reveal novel insights into the previously un(der)explored role of membrane tension in regulating viral fusion.
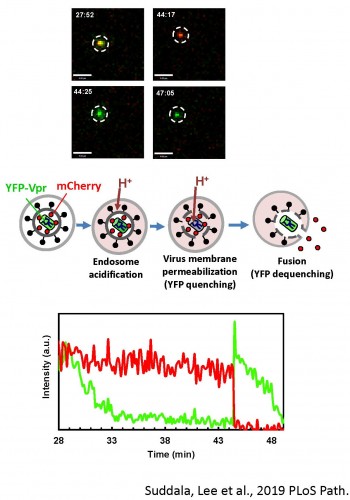
Old and New World arenaviruses can cause severe hemorrhagic fever in humans. There are currently no approved vaccines or drugs to battle arenavirus infection. Despite extensive research, arenavirus entry pathways into host cells and the mechanism of arenavirus glycoprotein (GPc) mediated virus-cell fusion are not well understood. We are optimizing single virus labeling and imaging tools to examine entry pathways and the fusion mechanism of arenaviruses, focusing on the Lassa virus. Our data suggest that arenavirus fusion is uniquely preceded by a viral membrane permeabilization step (Figure 5 and Movie 4).
http://www.ncbi.nlm.nih.gov/sites/myncbi/gregory.melikian.1/bibliography/40733965/public/?sort=date&direction=descending
Alumni: Currently at/Worked at:
Xiangyang Guo Senior Scientist, Biopharmaceutical company, Beijing China
Ashwanth Francis Assistant Professor, Florida State University
Satya Singh Postdoc, University of Massachusetts
Kristina Rowland Ipsum Diagnostics, Atlanta
Christine Lee Center for Disease Control, Atlanta
Junghwa Choi-Kirschman Postdoctoral Scientist, TGen
Caleb Mason Research Professional II, UGA
Chetan Sood NanoView BioSciences, Boston
Krishna Suddala Postdoc, NIH
Charline Giroud Postdoc, Oxford, UK
Tanay Desai Imaging Specialist, Carl Zeiss Microimaging
Jennifer Spence Research Assist. Prof., Albert Einstein, USA
Yoon-Hyeun Oum Staff Scientist, Emory, USA
Lusine Demirkhanyan Research Associate, University of Illinois, USA
Naoyuki Kondo Kansai Medical University, Japan
Kiran Verma Research Specialist, Emory, USA
Michelle de la Vega Research Scientist, Health Science Academy
Sergi Padilla-Parra Senior Lecturer, King’s College, London
Surbhi Jain Physician, USA
Rolf Suter Lab automation specialist, Switzerland
Nishi Sharma Postdoc, NIH, USA
Kosuke Miyauchi Scientist, RIKEN, Japan
Olga Latinovic Assistant Professor, University of Maryland, USA
Yuri Kim University of Maryland, USA
Grant Jones University of Maryland, USA
Gennadiy Novitskiy University of Maryland, USA
Naveen Jha FDA, USA
Patrick Millet University of North Carolina, USA
Victoria Saakian University of Maryland, USA
Vladimir Morozov Senior Scientist, Robert Koch Institute, Germany
Sergey Cheresiz Novosibirsk State University, Russia
Michael Leung University of Maryland College Park
Levon Abrahamyan Assistant Professor, University of Montreal, Canada
Visiting students/scholars/volunteers:
Nivriti Galhaut Atlanta, GA
David Gevorgyan Armenia
Pedro Matos UK
Margaret Black Atlanta, GA